Logical Statements
MolScript allows for precise subset selection based on a small number of logical commands. Combination of the proper logic calls and atom selections will allow for the drawing of any group of atoms or bond. The logical operators recognized by MolScript are:
- not
- and
- or
- require
- either
These logical operators can be combined into statements which make selections either positively (inclusive sets) or negatively (exclusive sets). Absolute discussion of the function of these paramaters is tedious and not as helpful examples applying them to common situations. Let's explore logical selection through two examples:
Atom selection for ball and stick
Showing residues on a ribbon diagram is often very instructive. Remember the standard syntax for drawing ball and stick representations in residue #:
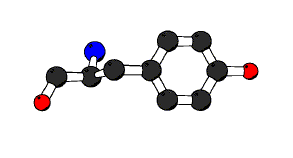
ball-and-stick in residue #;
This statement produces a ball and stick representation of all atoms in the residue indicated. It is often desired to only draw a limited set of atoms in a particular residue. In this case we'll draw just the sidechain of this tyrosine residue including the alpha carbon (atom CA):
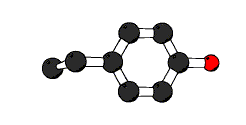
ball-and-stick require not either atom C, atom O or atom N and
in residue #;
In the above example the logical operators not, either, or,
and and are combined to prevent the drawing of three backbone
atoms, the carbonyl carbon, the carbonyl oxygen and the amide nitrogen.
These operates are concatenated into the same simple ball-and-stick command
from above. This is an example of a negative (exclusive) set. The statement
indicates (require) which atoms (either, or, and)
not (not) to draw. In the logic syntax it's important to note
the use of commas, spaces and the ever present semicolon.
The same output can be achieved with a different set of logical operators in a similar construct. This time the positive (inclusive) selection method will be used.

ball-and-stick require either atom CA, atom CB, atom CG, atom CD*,
atom CE*, atom CZ or atom OH and in residue #;
This time the operators are either, or, and and.
There are fewer operators used to write the positive statement, but for the
case of a tyrosine residue a much larger set of atom selections is required.
While not efficient for tyrosine, the positive statement is very efficient
when drawing only the sidechain of alanine (atom CB)! It is good to note the
use of the wildcard character (*) in the statement. The wildcard holds a
place for any other character. In a tyrosine residue there are two delta
carbon atoms, 1 & 2. Without the wildcard the selection,
atom CD* would have to be expanded to atom CD1,
atom CD2 to achieve the same results.
Atom selection in drawing bonds
The same logical operators apply in drawing bonds between specific atoms. Remember that the general form of the bonds command is:
bonds atom-selection atom-selection;
With this in mind, look at an example of the hydrogen bonds to aspartic acid 69 in E. coli HPr.
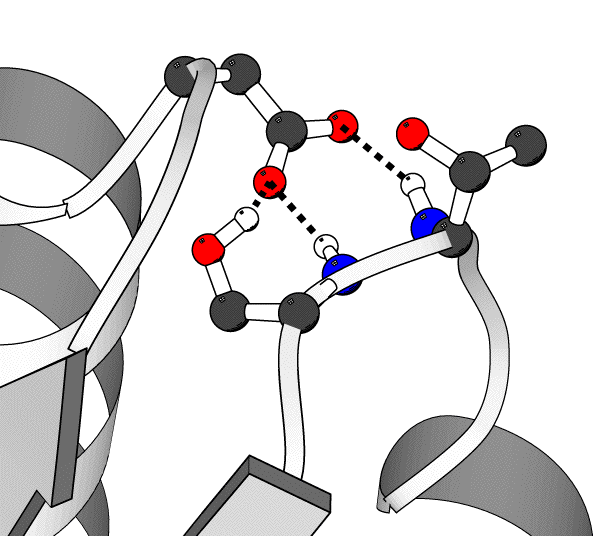
There are three hydrogen bonds to D69. They are drawn here as dashed lines. The bonds are between the hydroxyl proton of serine 31 and the OD1 atom of aspartic acid 69, the amide proton of serine 31 and the OD1 atom of aspartic acid 69, and the amide proton of threonine 30 and the OD2 atom of aspartic acid 69, donor to acceptor respectively. Each bond is drawn by a single command line in the Molscript script. The three lines use to generate these bonds are, in the same order as listed above in the text:
bonds require atom HG and in residue 31 require atom OD1 and in residue 69;
bonds require atom H and in residue 31 require atom OD1 and in residue 69;
bonds require atom H and in residue 30 require atom OD2 and in residue 69;
The general form for atom selection in the bonds command is
"atom type - in residue #." Each "atom-selection" runs from
require to the residue #. This meets the general form of
the command. With only one atom being specified, the require statement
is just require, no not and/or either.
The key to specifying the proper bonds is probably knowing the
crystallographic atom names. The parameters linewidth
and linedash were manipulated to produce the thick
dashed bonds.
Written and maintained by Kevin Shaw
Mail kevshaw at yea4us